Clinical trial search
Developing an aspect-based clinical trial search and an interpretable system using the Pubmed citation network
Publications and Awards
IEEE ICDH 2023 Full Paper that also got recognized as a “candidate for the Best Student Paper”. Received the Best Graduate Forum Presentation Award in COMSNETS 2018. One Full paper accepted in CSoNet 2019.
Proposal got accepted. Our clinical trial search work will now be further investigated as a new project titled “Developing a Fair and Interpretable Clinical Trial Search System” by Dr. Koustav Rudra (Project PI) of Centre of Excellence in Artificial Intelligence, IIT Kharagpur for the next 2 years.

Abstract
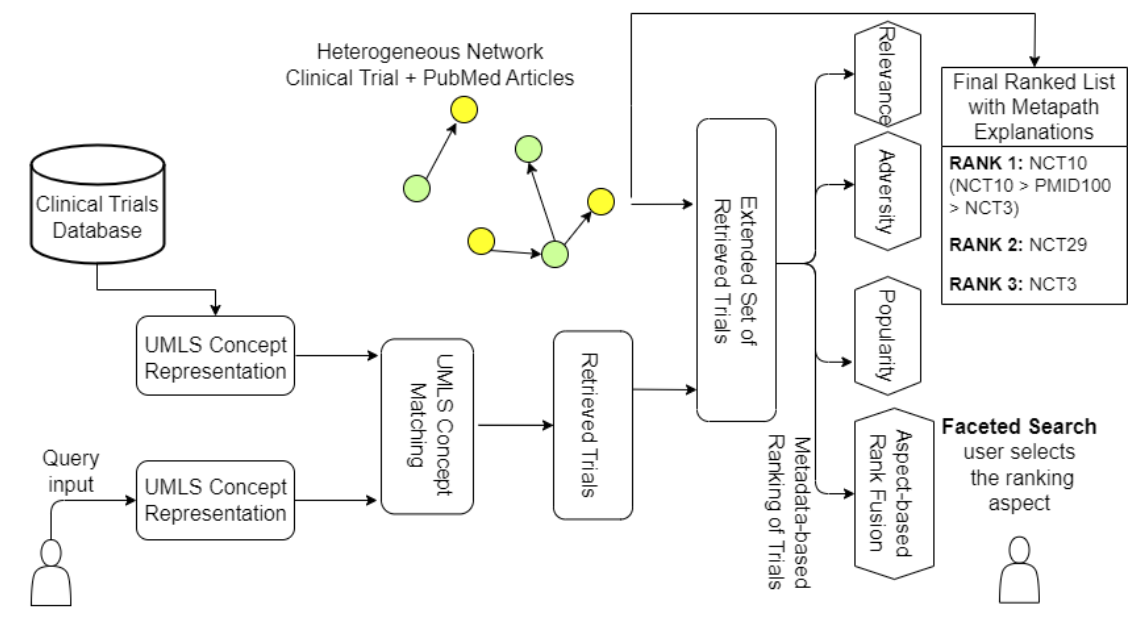
Clinical trials are an essential source of information for practicing Evidence-Based Medicine because they help to determine the efficacy of newly developed treatments and drugs. However, most of the existing trial search systems focus on a specific disease (e.g., cancer) and utilize disease-specific knowledge bases that hinder the adaptation of such methods to new diseases.
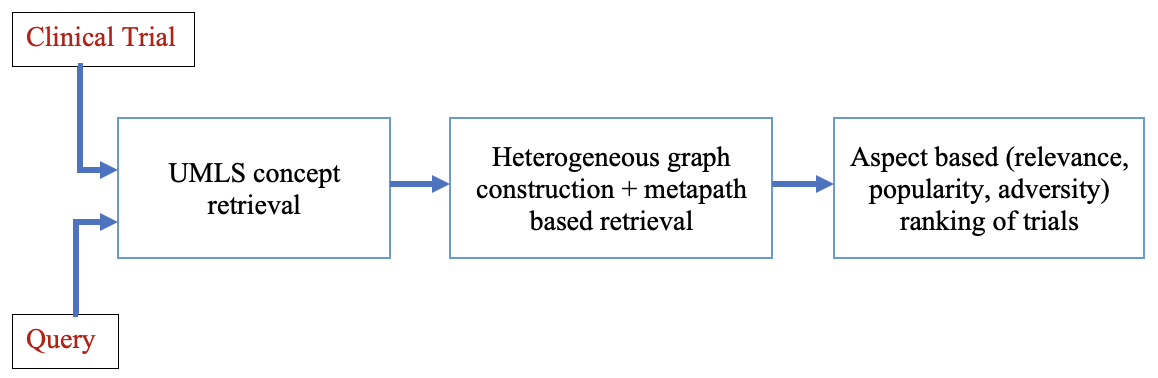
In this work, we overcome both limitations and propose a graph-based model that explores both clinical trials and the Pubmed databases to alleviate the shortage of relevant clinical trials for a query. We construct a large heterogeneous graph (750K nodes and 1.2 Million edges) made of clinical trials and Pubmed articles linked to clinical trials.
As both the graph edges and nodes are labeled, we develop a novel metapath-based similarity search (MPSS) method to retrieve and rank clinical trials across multiple disease classes. We primarily focus on consumers and users that do not have any prior medical knowledge.
As there are no multiple disease-wide trial search evaluation datasets, we contribute a high-quality, well-annotated query-relevant trial set comprising around 25 queries and, on average, approximately 95 annotated trials per query. We also perform a detailed evaluation of MPSS on the TREC Precision Medicine Benchmark Dataset, a disease-specific clinical trial search setting.